The "Human Jigsaw"
Resolving commingled skeletal assemblages is a major challenge in forensic anthropology as well as in bioarchaeology. In bioarchaeological contexts, multiple burials, typically leading to commingled remains from multiple individuals, have constituted a standard mortuary practice across the world throughout human history. Furthermore, the fragile nature of archaeological material usually results in fragmented skeletal remains, which makes sorting even more difficult to resolve. In forensic contexts, on the other hand, it is much more common to dealing with recently deposited human remains which are relatively better preserved and more often intact. Forensic cases with commingled remains may be due to criminal activity or it may concern deposition of bodies in mass graves as a result of warfare or a natural disaster. Despite that fact that there is an array of available methods ranging from standard measurements and meticulous visual observation to DNA fingerprinting and other biochemical process that can facilitate resolving commingled skeletal remains, these tend to overlap and most often complement one another in terms of efficiency, accuracy, suitability and available resources. Furthermore, in such forensic cases, there is also the time constraint whether due to media attention or legal issues. Hence, the very need for sorting methods that are fast, accurate but also cheap and easy to implement. This is a primary reason why research on osteometric sorting has attracted much attention over the past decade or so.
In general, osteometric sorting could be described as a way of quantifying bone morphology and using statistical models as a more reliable and efficient substitute for associating through visual observation. Up until recently, most osteometric approaches have relied either on standard measurements, which lack the capacity to adequately capture the morphology of the bones, or on full mesh-to-mesh comparisons in the 3D space, which due to statistical reasons provide marginally better performance than standard osteometric measurements. In any case, the Achilles' heel of osteometric sorting methods is their limited capacity to quantify bone morphology in the "same" way an experienced anthropologist makes an elaborate decision on pair-matching two bone antimeres by meticulously observing the bones' surface. Don't get this wrong! Modern 3D scanning technologies can capture a bone's complete morphology with great accuracy and at unprecedented quality, as you can also observe in our small 3D library section. It is just a matter of finding or inventing the "right" way to quantify it in a statistically useful manner for the osteometric sorting challenges at hand.

This is the methodological gap in osteometric sorting techniques that the "Human Jigsaw" project has been aiming to fill. The research work focuses on pair-matching the long bones of the lower limbs as strategy for individualizing femurs and tibias from commingled skeletal assemblages and mass burials. This is achieved with the utilization of novel morphometric traits that adequately describe the intrinsic morphology of the bones' surface and the development of robust methods to quantify them as well as the involvement of stochastic processes in the statistical analysis in order to maximize the performance of the proposed sorting algorithms. The osteometric sorting research of the "Human Jigsaw" project aims at two distinct but complementary pair matching strategies for the skeletal elements of the lower limbs. One strategy involves the pair matching of bilateral elements, as for example sorting the left and right femurs per individual. The other strategy targets the pair matching of adjacent elements, such as in sorting the left femur and left tibia per individual.
Between standard osteometric measurements and full scale 3D point cloud or mesh data, which are the commonly utilized in osteometric sorting as previously mentioned, the novelty in the "Human Jigsaw" project is that we devised an intermediate solution. The solution of morphometric traits! These are simple metrics that hold the statistical benefits of standard osteometric measurements (i.e. low dimensionality), yet they are elaborate descriptors of morphological shape variation, which can harvest the vast information available in a 3D mesh model of a bone's surface. To name a couple, the Imin/Imax ratio, which quantifies in a way how much elongated a diaphyseal cross section is, and the diaphyseal bending, which describes the amount of curvature along the main axis of the diaphysis. The idea behind this approach is to quantitatively capture the morphology to the long bone diaphysis, which would allow us to estimate the similarity between left and right side bone antimeres based on their anticipated symmetry within certain limits. For this reason, the CSG Toolkit, which was originally developed as a replacement for the laborious LCM, has been incorporated and further developed to calculate the additional morphometric traits required. As shown in the image below, the CSG Toolkit allows for automated yet accurate extraction of CSG properties and also provides us with additional morphometric traits for our osteometric sorting purposes.
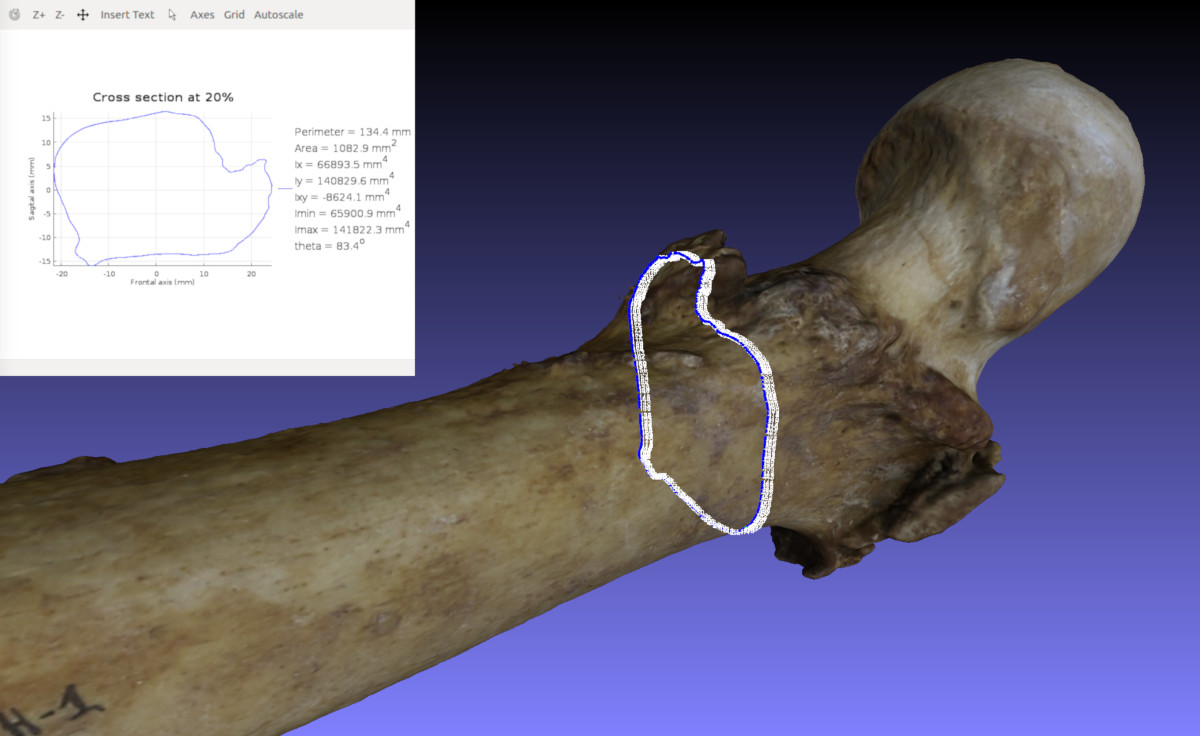
3D points (blue dots) of a cross-sectional contour of a femur shown in Meshlab and its corresponding 2D polyline and cross-sectional geometric properties calculated and plotted with the CSG Toolkit.
As said, a solid set of simple yet elaborate descriptors of the bone's morphology is a great start forward towards successful sorting of commingled bones. Once achieved, and the CSG Toolkit will do that in seconds given the availability of a 3D model, we still need a robust statistical approach to make good use of our freshly collected osteometric data, which brings us to the second novelty of the "Human Jigsaw" project. The usual statistical approaches to osteometric sorting have been either by inferring dissimilarity and hence exclusion (i.e. mismatched pair) based on the null hypothesis of symmetric bone antimeres or by evaluating similarity based on some distance metric such as Euclidean or even Hausdorff distance more recently as the most probable match. Common ground in all former approaches has been the merging of multivariate osteometric data into univariate statistical inference tests! Our sorting approach, although still dependent on descriptive statistics and sample distrubutions, has taken a completely different and novel approach with a multi-stage complex stochastic process involving multivariate descriptive statistics for exclusion of definite mismatches, Bayes' rule for conditional probability checking on plausible pairs and a modified k-NN searching algorithm to facilitate the optimal sorting of bone antimeres in a given assemblage. More details on the sorting algorithm can be found in the original publication "Advances in Osteometric Sorting: Utilizing Diaphyseal CSG Properties for Lower Limb Skeletal Pair-Matching".
Paramount for any newly proposed method, no matter how novel, elaborate, accurate or even fancy it may be, is its efficiency in terms of applicability and ease of use. Apparently, osteometric sorting methods are no exception to this. Hence, the newly proposed sorting method has been implemented in single simple to use GNU Octave function named sorting_BE licensed under the GPLv3. Although sorting_BE does not have a GUI and can only be invoked from the GNU Octave environment, the figure below illustrates the results of successful pair matching performed on ten femurs belonging to five individuals from the Athens Collection. The figure includes only two out of the five cross sections utilized for pair matching. Nevertheless, careful observation between the matched pairs and in contrast to the all other available samples shall reveal the symmetric relation between matching pairs. These similarities/dissimilarities, heavily bound to the intrinsic morphology of each and every bone, allow well-trained eye of a professional examiner to qualitatively sort commingled bones but up until now have been so difficult for osteometric methods to quantify.
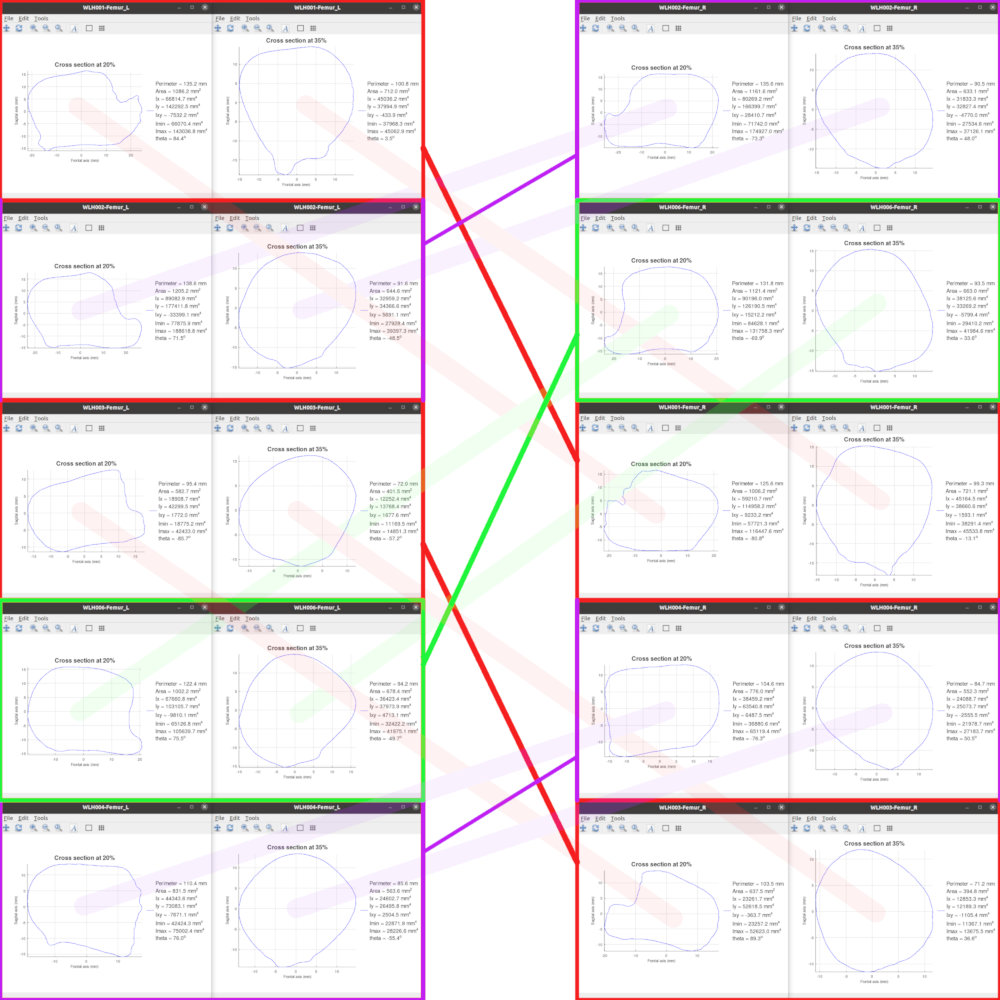
The matching pairs (indicated by colored lines) resulted from the analysis of an assemblage of left and right femurs of five individuals. On the left-hand and right-hand side of the image are two of the five cross-sectional contour plots along with the related CSG properties of the left and right femurs, respectively
The sorting_BE function has been repeatedly tested with assemblages of up to 220 bones randomly selected from the available pool of femurs and tibias form the Athens Collection and its underlying algorithm has demonstrated unprecedented sorting accuracy (~99% up to 100%) in various tested scenarios. More over, the implemented pair matching algorithm can successfully handle cases with large disparity (i.e. commingled assemblages with a large number of single bones without a matching pair) since it is capable of correctly identifying also single elements when they do not have a matching pair in the commingled assemblage. The ancient reducio ad absurdum embedded in a modern statistical approach to facilitate a long standing but still pressing problem:
Sorting commingled skeletal remains!
The "Human Jigsaw" project was funded by the European Union's Horizon 2020 research and innovation program under the Marie Skłodowska-Curie Actions granted to Maria-Eleni Chovalopoulou [Program/Call: H2020-MSCA-WF-2018, Proposal: 867438-Human Jigsaw]. It was conducted at the STARC of The Cyprus Institute under the supervision of Ass. Prof. Efthymia Nikita and in collaboration with Andreas Bertsatos, researcher at the NKUA.

